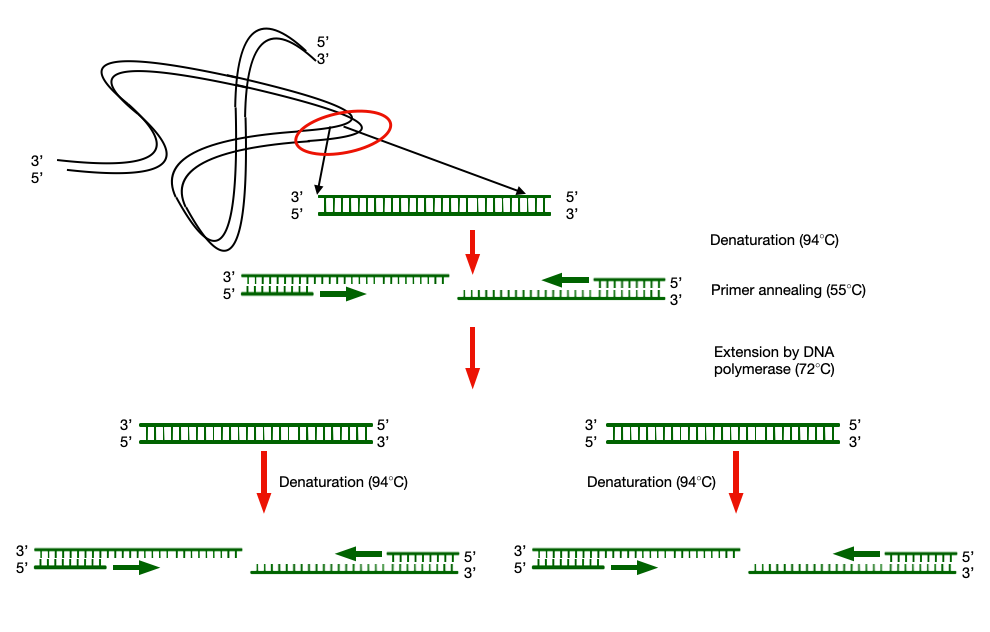
Principle of Polymerase chain reaction (PCR)
This method is analogous to in-vivo DNA replication as it uses DNA polymerase which catalyses the polymerisation at the free 3’OH end of the primer bound to the template DNA. A pair of primer is required to amplify the target sequence on two complementary strands of the template DNA.
Definition of PCR
A molecular laboratory technique that uses an entangled Template DNA to produce multiple copies of the desired short DNA fragment by using a pair of DNA primer, DNA polymerase and template DNA.
History of PCR
The PCR which stands for Polymerase Chain Reaction, is developed by Kary Mullis in the 1980s. For every revolutionary scientific method, there is a breakthrough discovery which opens newer avenues for advanced experimentation. In 1950s, Arthur Kornberg found out the most sought enzyme at that time from E.coli, now known as DNA polymerase. This discovery was a real breakthrough in molecular research and later formed a basis for in-vitro DNA polymerisation. We have read about a decade long journey of the discovery of DNA polymerase in the article Arthur Kornberg: A doctor who discovered a knitting enzyme.
But again, there is no success without hardships and struggle. Difficulties occurring in the Sanger sequencing method was a major cause behind the discovery of PCR. Sanger sequencing was being used for identification of the gene sequence at that time and for the detection of the allelic differences between the DNA samples. However, it was very frustrating to get the correct sequence without spending huge amount of resources, efforts and patience. The reason is that the Sanger sequencing could not yield stronger signals due to the fact that the DNA to be sequenced could not be made available in sufficient quantity.
Mullis wanted to develop some new and faster method so that he will be able create multiple copies of the DNA fragment to be sequenced correctly but he had no idea how to go about it. He wanted to amplify specific fragment from a given DNA sequence. To facilitate this, he came up with a trick to use primers that will bind to both complementary strands of the template DNA.
One primer will bind at the starting point of the desired sequence (Forward primer) and the other will bind to the end of the desired sequence to be amplified and copy the corresponding template strands into two new DNA products on every binding of DNA polymerase to the DNA template. The Product of one reaction was now available for the next round of copying, thus initiating a chain of replicating reaction and hence the name Polymerase Chain Reaction (PCR). This sounded so miraculous and relieving, but it was not as easy as it sounded.
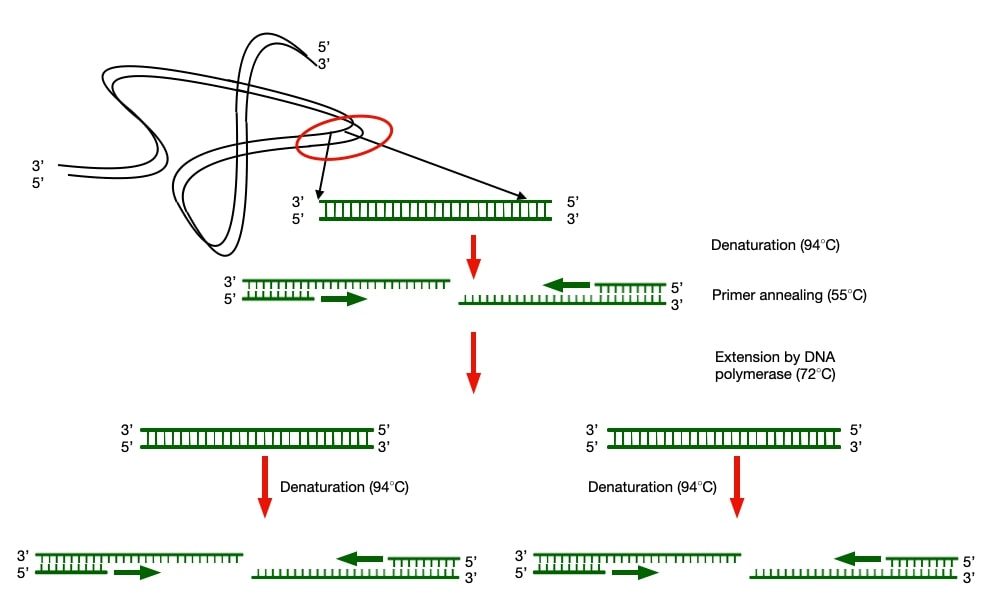
He started off by using an E.coli DNA polymerase for the amplification of the DNA fragment. As the reaction was being carried out in an in-vitro condition, there was not any enzyme which will separate each strand of the template DNA for subsequent binding of the primers and DNA polymerase. Therefore, he had to heat the samples at the denaturing temperature so that hydrogen bonds between two strands will break.
Unfortunately, this came with a major disadvantage as the denaturing temperature could destroy the functional DNA polymerase. Therefore, he had to add a fresh enzyme extract to continue the DNA amplification. This solved the important purpose of amplifying any desired DNA fragment at a required amount in laboratory. But, it was very difficult to conduct the PCR due to lack of automation and availability of the enzyme which could be stable at a high temperature.
Ultimately, the first thermostable enzyme, which is now popular as Taq DNA polymerase, was isolated from the thermophilic bacteria known as Thermus aquaticus. Taq DNA polymerase was able to withstand the denaturing temperature of the reaction even after multiple cycles of the reactions. With the constant improvement and modification in the automation of the instrument, most sophisticated PCR machines are available today.
Components of PCR
- Template DNA:
It can be any DNA sample containing a short desired DNA sequence of interest which is required to be amplified.
- Primers:
DNA primers are small single stranded chain of nucleotides (oligonucleotides) complementary to the target DNA sequence. Primers bind to the template DNA and prime the extension by DNA polymerase.
- DNA polymerase:
It is an enzyme required for the extension at 3’OH end of the primer bound to the Template DNA.
- dNTPs (deoxyribonucleotides):
These are building blocks used by DNA polymerase to extend the primers leading to the synthesis of the new complementary DNA strand to the template DNA. These nucleotides also provide the energy required for the the DNA synthesis.
- Magnesium:
Being positively charged (Mg2+), Magnesium helps to stabilise negatively charged dNTPs during the interaction between DNA polymerase and template DNA strand.
Applications of PCR:
Earlier, this technique was being used for limited purposes only, however, in today’s time it offers innumerable applications in various areas. Some of these areas are listed below
- Gene probe labelling
- DNA fragment amplification
- DNA sequencing
- Quantification of RNA- RT PCR
- Biotechnological applications
Simplicity and reproducibility of PCR has popularised this method to every biology laboratory. Combining it with bioinformatics tools, it will open newer areas of research in the coming times. Try out some exercise related to real time PCR here.
Read more about PCR